Abstract
Introduction: Recent genome-wide studies of leukemia have revealed its clonal architecture and subclonal heterogeneity. Patient-derived xenograft (PDX) models are assumed to capture the cellular and molecular characteristics of human cancer and widely utilized for studying drug response. However, it has not been elucidated whether PDXs faithfully represent the genetic features of primary leukemia. In this study, we monitored the dynamics of somatic mutations in primary leukemia cells and PDX cells to evaluate their genetic stability and clonal selections.
Methods: A total of 90 fresh bone marrow or peripheral blood samples from patients with hematological malignancies were intravenously transplanted into NOD/Shi-scid, IL-2Rγnull (NOG) mice at 0.2-15x106 cells per mouse. Targeted sequencing of 54 genes frequently identified in myeloid malignancies was performed in 40 paired genomic DNA of successfully engrafted AML patient and their PDX samples and 20 AML patients with engraftment failure. Clonal diversities were analyzed by comparing variant allele frequencies (VAF) of somatic mutations in 16 sets of serial samples from AML patients at diagnosis and subsequent relapse, and their PDX models. All patients provided written informed consent.
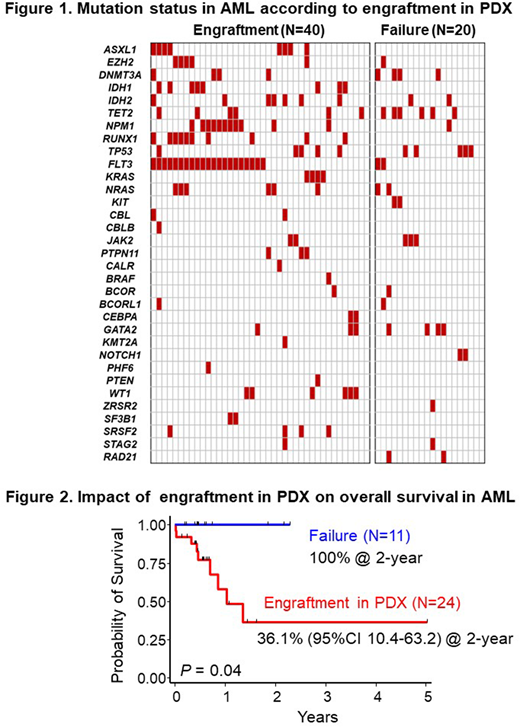
Results: Sixty-five of 90 (72%) primary cells (AML; 60, ALL; 5) engrafted in NOG mice at the median of day 112 (29-549) post transplantation. Patient samples with successful establishment of PDX models included significantly higher number of infused cell counts (7.2 vs. 4.9 x 106 cells, P=0.002), higher percentage of blasts (56.2% vs. 38.7%, P=0.04) and harbored activating kinase gene mutations more frequently than those with engraftment failure (83% vs. 40%, P=0.001; Figure 1). Especially, FLT3 (53% vs. 10%, P=0.001), NPM1 (28% vs. 5%, P=0.04), RUNX1 (25% vs. 0%, P=0.01), IDH1 (20% vs. 0%, P=0.03) and ASXL1 (18% vs. 0%, P=0.05) mutations were significantly accumulated in successfully engrafted patients. In terms of the sensitivity to chemotherapy, patients with successful engraftment in NOG mice showed poorer 2-year overall survival than those without engraftment (36.1% vs. 100%, P=0.04; Figure 2).
In 40 pairs of AML patients and their PDXs, a total of 172 genetic alterations (101 SNV, 39 frameshift, 32 inframe insertion/deletion) were identified. Mean VAF changes in PDXs compared with patients showed dominant elevation in driver gene mutations including FLT3 (+16.9%), CEBPA (+19.1%) and WT1 (+26.4%) demonstrating the expansion of clones carrying these mutations in PDXs, whereas those of ASXL1 (-2.2%), NPM1 (+4.8%) and DNMT3A (+6.0%) in PDXs were almost the same as primary patient samples, implicating that these mutations are founder events in clonal evolution. VAF of 26 pairs of patients' samples and their PDXs at relapse were more concordant than those of 12 pairs at diagnosis (r2=0.689 vs. 0.550).
In 16 sets of AML patients' samples at diagnosis and relapse, and their PDX samples, clonal diversities between primary leukemia at diagnosis and their PDXs were classified into 2 subtypes. In 7/16 models (44%), minor clones detected in patients at diagnosis dominantly expanded to be major clones in their PDX models, whereas major clones persisted in similar VAF between primary leukemia and their PDXs in 9/16 models (56%). These subtypes of clonal diversity between primary leukemia at diagnosis and their PDXs were concordant with clonal evolution between patients at diagnosis and relapse in 12/16 models (75%). Furthermore, in 4/16 models (25%), clones selectively expanded in PDXs were detected as major clones in their patients' first relapsed samples predictively.
Conclusions: Our findings indicate the existence of diverse clonal evolution in AML PDX models. Primary AML samples harboring treatment-resistant clones have higher potential of engraftment and growth in NOG mice. Furthermore, PDX models recapitulate the clonal evolution from diagnosis through relapse in treatment-resistant patients. Further understanding of expanded clones in PDX models could reveal the pathogenesis of clonal selection in AML patients.
Kiyoi:Bristol-Myers Squibb: Honoraria; FUJIFILM Corporation: Research Funding; Zenyaku Kogyo Co., Ltd.: Research Funding; Sanofi K.K.: Research Funding; Celgene Corporation: Research Funding; Sumitomo Dainippon Pharma Co., Ltd.: Research Funding; Phizer Japan Inc.: Research Funding; Kyowa Hakko Kirin Co., Ltd.: Research Funding; Eisai Co., Ltd.: Research Funding; Astellas Pharma Inc.: Research Funding; Otsuka Pharmaceutical Co., Ltd.: Research Funding; Novartis Pharma K.K.: Research Funding; Takeda Pharmaceutical Co., Ltd.: Research Funding; Nippon Shinyaku Co., Ltd.: Research Funding; Chugai Pharmaceutical Co., Ltd.: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal